TopP–S: Persistent homology‐based multi‐task deep neural networks for simultaneous predictions of partition coefficient and aqueous solubility
Kedi Wu; Zhixiong Zhao; Renxiao Wang; Guo‐Wei Wei
Index: 10.1002/jcc.25213
Full Text: HTML
Abstract
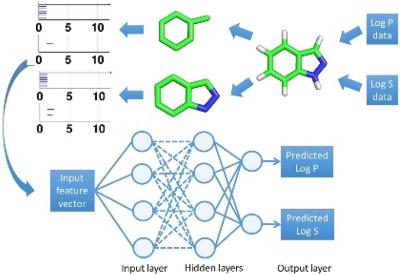
Aqueous solubility and partition coefficient are important physical properties of small molecules. Accurate theoretical prediction of aqueous solubility and partition coefficient plays an important role in drug design and discovery. The prediction accuracy depends crucially on molecular descriptors which are typically derived from a theoretical understanding of the chemistry and physics of small molecules. This work introduces an algebraic topology‐based method, called element‐specific persistent homology (ESPH), as a new representation of small molecules that is entirely different from conventional chemical and/or physical representations. ESPH describes molecular properties in terms of multiscale and multicomponent topological invariants. Such topological representation is systematical, comprehensive, and scalable with respect to molecular size and composition variations. However, it cannot be literally translated into a physical interpretation. Fortunately, it is readily suitable for machine learning methods, rendering topological learning algorithms. Due to the inherent correlation between solubility and partition coefficient, a uniform ESPH representation is developed for both properties, which facilitates multi‐task deep neural networks for their simultaneous predictions. This strategy leads to a more accurate prediction of relatively small datasets. A total of six datasets is considered in this work to validate the proposed topological and multitask deep learning approaches. It is demonstrated that the proposed approaches achieve some of the most accurate predictions of aqueous solubility and partition coefficient. Our software is available online at http://weilab.math.msu.edu/TopP-S/. © 2018 Wiley Periodicals, Inc.
|
TopoMS: Comprehensive topological exploration for molecular ...
2018-03-23 [10.1002/jcc.25181] |
|
Cover Image, Volume 39, Issue 12
2018-03-15 [10.1002/jcc.25210] |
|
Dispersion interactions between neighboring Bi atoms in (BiH...
2018-03-13 [10.1002/jcc.25209] |
|
Can we predict the structure and stability of molecular crys...
2018-03-12 [10.1002/jcc.25198] |
|
Fast and flexible gpu accelerated binding free energy calcul...
2018-03-12 [10.1002/jcc.25187] |