| Structure | Name/CAS No. | Articles |
|---|---|---|
 |
Nuclease S1
CAS:37288-25-8 |
|
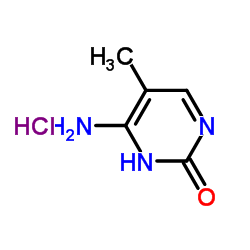 |
5-METHYLCYTOSINE HYDROCHLORIDE
CAS:58366-64-6 |
|
 |
Nuclease P1
CAS:54576-84-0 |
|
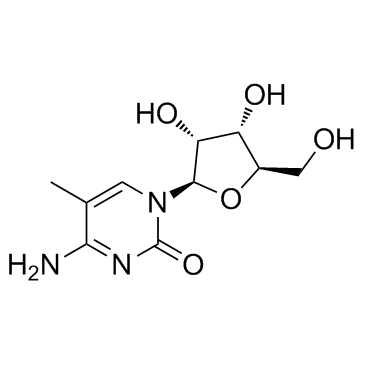 |
5-Methylcytidine
CAS:2140-61-6 |