Ribosomal binding and dipeptide formation by misacylated tRNA(Phe),S.
T G Heckler, J R Roesser, C Xu, P I Chang, S M Hecht
Index: Biochemistry 27 , 7254-7262, (1988)
Full Text: HTML
Abstract
Eight structurally modified peptidyl-tRNA(Phe),s were employed to study P-site binding and peptide bond formation in a cell-free system involving Escherichia coli ribosomes programmed with poly(uridylic acid). It was found that the two analogues (N-acetyl-D-phenylalanyl-tRNA(Phe) and N-acetyl-D-tyrosyl-tRNA(Phe] containing D-amino acids functioned poorly as donors in the peptidyltransferase reaction and that two N-acetyl-L-phenylalanyl-tRNA(Phe)'s differing from the prototype substrate in that they contained 2'- or 3'-deoxyadenosine at the 3'-terminus failed to form dipeptide at all when L-phenylalanyl-tRNA(Phe) was the acceptor tRNA. Interestingly, all four of these peptidyl-tRNA's bound to ribosomes to about the same extent as tRNA's that functioned normally as donors in the peptidyltransferase reaction, at least in the absence of competing peptidyl-tRNA species. Two peptidyl-tRNA's lacking an amino group were also tested. In comparison with N-acetyl-L-phenylalanyl-tRNA(Phe) it was found that trans-cinnamyl-tRNA(Phe) and 3-phenylpropionyl-tRNA(Phe)'s formed dipeptides to the extent of 53 and 80%, respectively, when L-phenylalanyl-tRNA(Phe)was used as the acceptor tRNA. N-Acetyl-beta-phenylalanyl-tRNA(Phe) was found to be the most efficient donor substrate studied. Both isomers transferred N-acetyl-beta-phenylalanine to L-phenylalanyl-tRNA(Phe); the nature of the dipeptides formed in each case was verified by HPLC in comparison with authentic synthetic samples. Further, the rate and extent of peptide bond formation in each case exceeded that observed with the control tRNA, N-acetyl-L-phenylalanyl-tRNA(Phe).
Related Compounds
| Structure | Name/CAS No. | Molecular Formula | Articles |
|---|---|---|---|
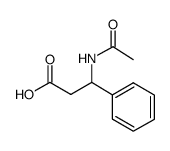 |
Ac-DL-β-Phe-OH
CAS:40638-98-0 |
C11H13NO3 |
|
Biodegradation of high molecular weight lignin under sulfate...
2009-02-01 [Bioresour. Technol. 100 , 1622-1627, (2009)] |
|
RNAi-mediated suppression of p-coumaroyl-CoA 3'-hydroxylase ...
2008-03-18 [Proc. Natl. Acad. Sci. U. S. A. 105 , 4501-4506, (2008)] |
|
Mutations in the cinnamate 4-hydroxylase gene impact metabol...
2009-12-01 [Plant J. 60 , 771-782, (2009)] |
|
A simple method to determine trypsin and chymotrypsin inhibi...
2004-06-30 [J. Biochem. Biophys. Methods 59 , 241-251, (2004)] |
|
Chemotactic activity from rabbit peritoneal neutrophils. Lac...
1976-08-12 [Biochim. Biophys. Acta 445 , 112-117, (1976)] |