| Structure | Name/CAS No. | Articles |
|---|---|---|
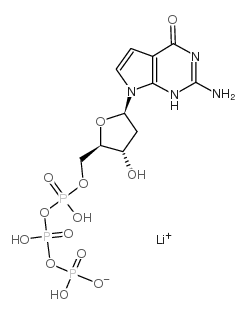 |
7-Deaza-2′-deoxyguanosine 5′-triphosphate
CAS:101515-08-6 |
|
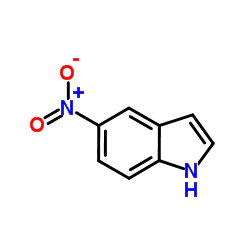 |
5-Nitroindole
CAS:6146-52-7 |