Bleomycin and X-ray-hypersensitive Chinese hamster ovary cell mutants: genetic analysis and cross-resistance to neocarzinostatin.
C N Robson, A Hall, A L Harris, I D Hickson
Index: Mutat. Res. 193 , 157-165, (1988)
Full Text: HTML
Abstract
We have previously reported the isolation of 3 mutants of Chinese hamster ovary cells which exhibit hypersensitivity to bleomycin. 2 mutants were isolated on the basis of bleomycin-sensitivity [designated BLM-1 and BLM-2, Robson et al., Cancer Res., 45 (1985) 5304-5309] and 1 as adriamycin-sensitive [ADR-1, Robson et al., Cancer Res., 47 (1987) 1560-1565]. Because bleomycin generates DNA-strand breaks via a free-radical mechanism, we have studied the survival response of these mutants to a range of drugs which also generate free radicals and consequently DNA-strand breaks. The mutants are all hypersensitive to phleomycin, which differs from bleomycin in being unable to intercalate due to a modified bithiazole moiety. However, BLM-2 cells alone are hypersensitive to pepleomycin, a semi-synthetic bleomycin analogue. In contrast, BLM-1 cells are more sensitive than BLM-2 to streptonigrin (which operates via a hydroquinone intermediate). ADR-1 cells show wild-type resistance to streptonigrin. The results obtained with neocarzinostatin, an antibiotic requiring thiol activation, are unusual in that both BLM-1 and BLM-2 are approximately 3-fold more resistant than parental cells. However, the steady-state intracellular level of the major non-protein thiol, glutathione, is not altered in BLM-1 or BLM-2 cells. ADR-1 cells show essentially wild-type resistance to neocarzinostatin. Analysis of cell hybrids shows that BLM-1 and BLM-2 cells are phenotypically recessive in combination with parental CHO-K1 cells and represent different genetic complementation groups not only from one another, but also from the bleomycin-sensitive mutant xrs-6, isolated on the basis of X-ray sensitivity by Jeggo and Kemp [Mutation Res., 112 (1983) 313-319]. These results indicate that at least 3 gene products are involved in cellular protection against bleomycin toxicity in mammalian cells.
Related Compounds
| Structure | Name/CAS No. | Molecular Formula | Articles |
|---|---|---|---|
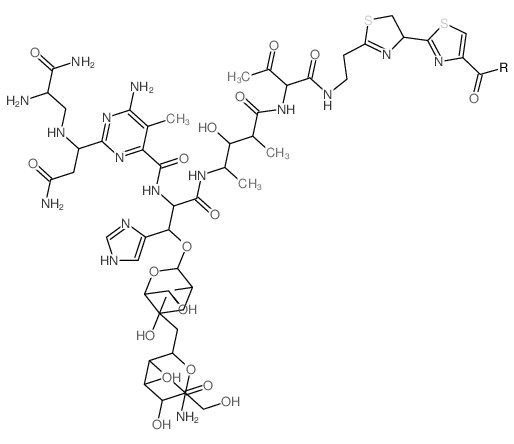 |
Phleomycin
CAS:11006-33-0 |
C52H75N16O21S2 |
|
CRE recombinase-based positive-negative selection systems fo...
2008-01-01 [Mol. Biochem. Parasitol. 157(1) , 73-82, (2008)] |
|
Copper(II).bleomycin, iron(III).bleomycin, and copper(II).ph...
1981-02-03 [Biochemistry 20 , 665-671, (1981)] |
|
Sulphydryl-mediated DNA breakage by phlemomycin in Escherich...
1977-02-01 [Mutat. Res. 42 , 181-190, (1977)] |
|
A Saccharomyces cerevisiae phleomycin-sensitive mutant, ph14...
1996-12-01 [Can. J. Microbiol. 42 , 1263-1266, (1996)] |
|
Extending the Schizosaccharomyces pombe molecular genetic to...
2014-01-01 [PLoS ONE 9(5) , e97683, (2014)] |