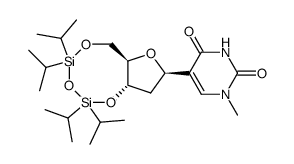
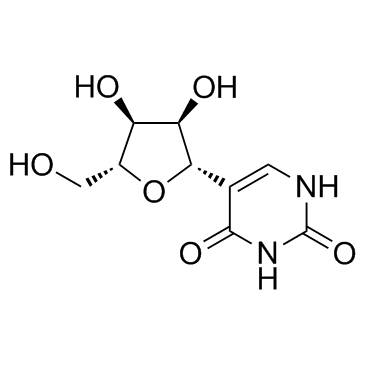
65358-15-8
| Name | pseudothymidine |
|---|---|
| Synonyms |
N-Methyl-1-phenylethylenediamine
N1-methyl-1-phenyl-ethanediyldiamine 2-Amino-1-methylamino-1-phenyl-aethan 1-methyl-2'-deoxypseudouridine N1-methyl-1-phenylethylenediamine N1-Methyl-1-phenyl-aethandiyldiamin N1-Methyl-1-phenyl-aethylendiamin N1-methyl-1-phenyl-1,2-ethanediamine N1-metylo-2-fenyloetylenodiamina 2'-deoxy-1-methylpseudouridine |
| Description | Pseudothymidine is a C-nucleoside analog of thymidine. |
|---|---|
| Related Catalog | |
| In Vitro | Pseudothymidine is a C-nucleoside analog of thymidine[1]. The calculated ΔΔG°50/mod is -0.5 kcal/mol, with a ΔTm/mod of 0.82°C. For the duplexes containing nine dA-T/ψT pairs, the ΔTm/mod is -0.9°C and a ΔΔG°50/mod is +1.1 kcal/mol. The modification of the duplex containing 12 consecutive dA-T/ψT base pairs produces a ΔTm/mod of -0.9°C and a ΔΔG°50/mod of +1.2 kcal/mol[2]. |
| Kinase Assay | Thermal DNA duplex denaturation studies are performed with templates containing up to twelve consecutive dA residues that are paired with its complement template containing consecutive T or Pseudothymidine (ψT) residues. Experiments are performed in a buffer (45 mM NaCl, 45 mM sodium citrate, pH 8.1, final vol. 1.5 mL) containing template and its complement (1.5 μM of each). Absorbance (260 nm) is monitored over a range of 25.0 to 90.0°C with a change in temperature of 0.5°C/min for five heating cycles. The initial heating cycle is discarded and the Tm is determined by averaging the temperatures of the remaining four cycles. The ΔTm between similar duplexes is calculated by subtracting the Tm of the duplex containing standard bases from the Tm of the duplex containing C-glycosides (including Pseudothymidine)[2]. |
| References |
| Molecular Formula | C10H14N2O5 |
|---|---|
| Molecular Weight | 242.22900 |
| Exact Mass | 242.09000 |
| PSA | 115.41000 |
| Storage condition | 2-8℃ |
| Precursor 4 | |
|---|---|
| DownStream 0 | |