| 结构式 | 名称/CAS号 | 全部文献 |
|---|---|---|
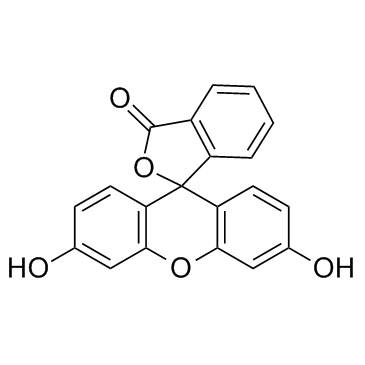 |
荧光素
CAS:2321-07-5 |
|
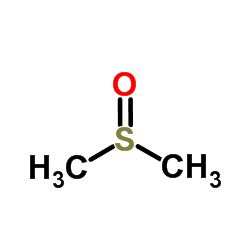 |
二甲基亚砜
CAS:67-68-5 |
|
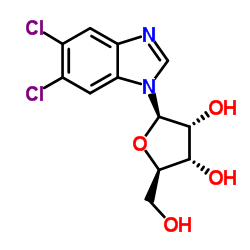 |
二氯苯并咪唑呋喃型核糖苷
CAS:53-85-0 |
|
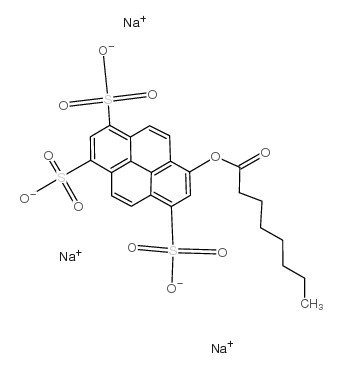 |
8-辛酰氧基芘-1,3,6-三磺酸三钠盐
CAS:115787-84-3 |