| Structure | Name/CAS No. | Articles |
|---|---|---|
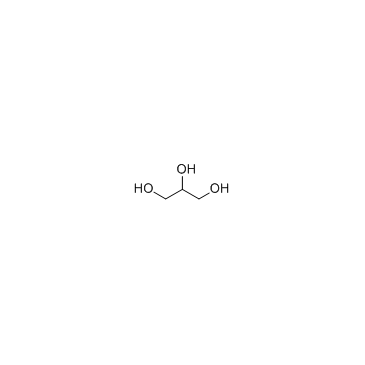 |
Glycerol
CAS:56-81-5 |
|
 |
Hydrochloric acid
CAS:7647-01-0 |
|
 |
sodium chloride
CAS:7647-14-5 |
|
 |
L-(+)-Lysine monohydrochloride
CAS:657-27-2 |
|
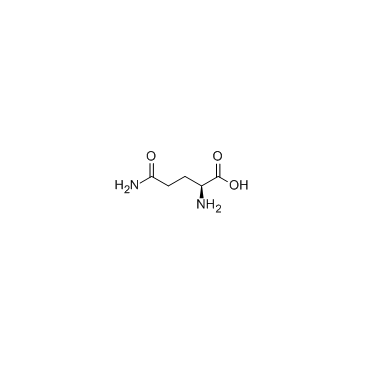 |
L-Glutamine
CAS:56-85-9 |
|
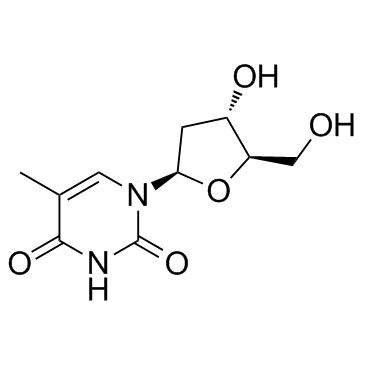 |
Thymidine
CAS:50-89-5 |
|
 |
SODIUM CHLORIDE-35 CL
CAS:20510-55-8 |
|
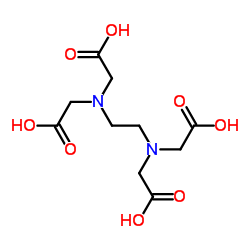 |
Ethylenediaminetetraacetic acid
CAS:60-00-4 |
|
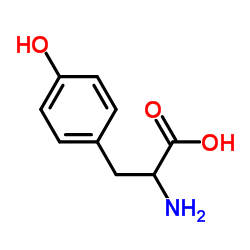 |
DL-Tyrosine
CAS:556-03-6 |
|
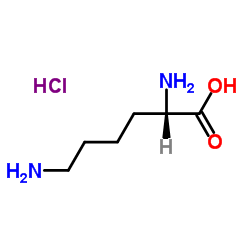 |
L-Lysine hydrochloride
CAS:10098-89-2 |