| Structure | Name/CAS No. | Articles |
|---|---|---|
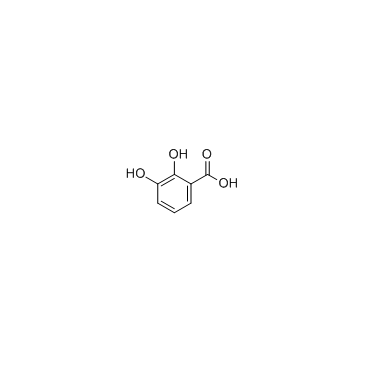 |
2,3-Dihydroxybenzoic acid
CAS:303-38-8 |
|
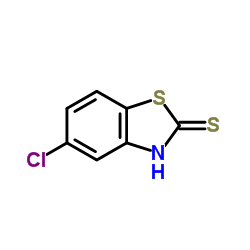 |
2-metcapto-5-chloro-benzothiazole
CAS:5331-91-9 |
|
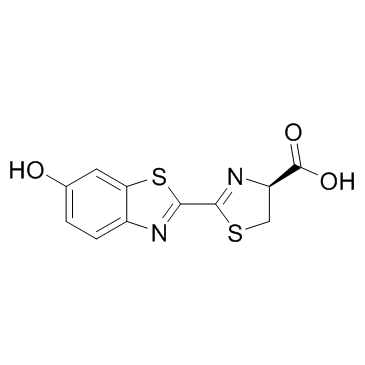 |
D-Luciferin
CAS:2591-17-5 |