| Structure | Name/CAS No. | Articles |
|---|---|---|
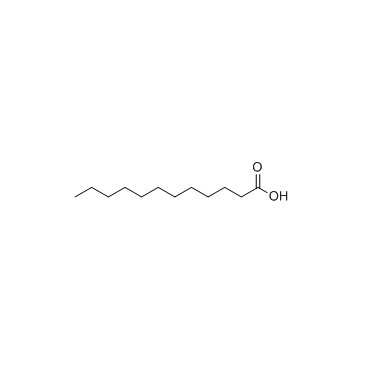 |
Lauric acid
CAS:143-07-7 |
|
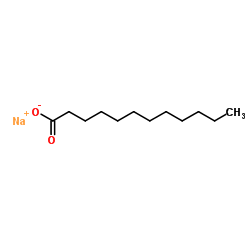 |
Sodium laurate
CAS:629-25-4 |
|
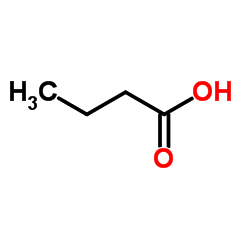 |
Butyric Acid
CAS:107-92-6 |
|
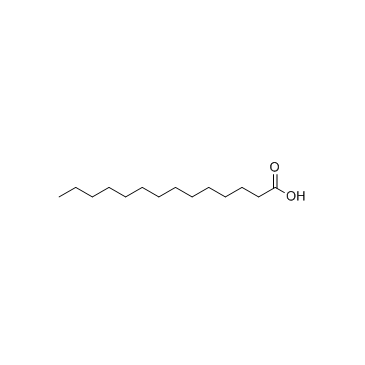 |
Myristic acid
CAS:544-63-8 |
|
 |
Lipoteichoic Acid
CAS:56411-57-5 |