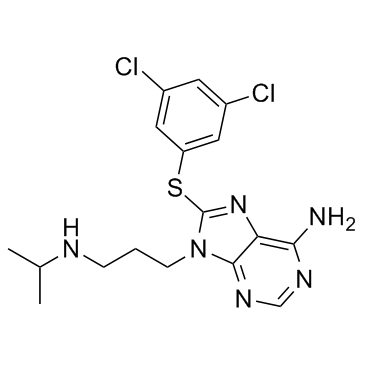
1454619-14-7
| Name | pu-ws13 |
|---|---|
| Synonyms |
8-(3,5-dichlorophenylthio)-9-(3-(isopropylamino)propyl)-9H-purin-6-amine
8-((3,5-Dichlorophenyl)thio)-9-(3-(isopropylamino)propyl)-9H-purin-6-amine 8-[(3,5-Dichlorophenyl)sulfanyl]-9-[3-(isopropylamino)propyl]-9H-purin-6-amine |
| Description | PU-WS13 is a selective Grp94 inhibitor, with an EC50 of 0.22 μM. |
|---|---|
| Related Catalog | |
| Target |
GRP94:0.22 μM (EC50) HSP90α:27.3 μM (EC50) HSP90β:41.8 μM (EC50) TRAP-1:7.3 μM (EC50) |
| In Vitro | PU-WS13 is a Grp94 inhibitor, with an EC50 of 0.22 μM. PU-WS13 also slightly suppresses Hsp90α, Hsp90β and Trap-1, with EC50s of 27.3, 41.8 and 7.3 μM, respectively. PU-WS13 (2.5-20 μM) shows no toxicity on two nonmalignant cell lines. PU-WS13 (15 μM) disrupts the circular architecture of HER2 at the plasma membrane of SKBr3 cells mediated through Grp94. PU-WS13 inhibits Grp94, and the inhibition induces apoptosis in and reduce the viability of HER2 overexpressing breast cancer cells[1]. |
| Kinase Assay | The Hsp90 FP competition assays are carried out in black 96-well micro-plates in a total volume of 100 μL in each well. A stock of 10 μM cy3B-GM and PU-FITC3 is prepared in DMSO and diluted with Felts buffer (20 mM Hepes (K), pH 7.3, 50 mM KCl, 2 mM DTT, 5 mM MgCl2, 20 mM Na2MoO4 and 0.01% NP40 with 0.1 mg/mL BGG). To each well is added the fluorescent dye-labeled Hsp90 ligand (6 nM cy3B-GM for Hsp90α, Hsp90β and Grp94 and 3 nM PU-FITC3 for Trap-1), protein (10 nM Hsp90α, 10 nM Hsp90β, 10 nM Grp94, 30 nM Trap-1) and tested inhibitor (including PU-WS13, initial stock in DMSO) in a final volume of 100 μL Felts buffer. Compounds are added in duplicate or triplicate wells. For each assay, background wells (buffer only), tracer controls (free, fluorescent dye-labeled Hsp90 ligand only) and bound controls (fluorescent dye-labeled Hsp90 ligand in the presence of protein) are included on each assay plate. The assay plate is incubated on a shaker at 4°C for 24 h, and the FP values (in mP) are measured. The fraction of fluorescent dye-labeled Hsp90 ligand bound to Hsp90 is correlated to the mP value and plotted against values of competitor concentrations. The inhibitor concentration at which 50% of bound fluorescent dye-labeled Hsp90 ligand is displaced is obtained by fitting the data. For cy3B-GM, an excitation filter at 530 nm and an emission filter at 580 nm are used with a dichroic mirror of 561 nm. For PU-FITC3, an excitation filter at 485 nm and an emission filter at 530 nm are used with a dichroic mirror of 505 nm. All of the experimental data are analyzed, and binding affinity values are given as relative binding affinity values (EC50, concentration at which 50% of fluorescent ligand is competed off by compound)[1]. |
| Cell Assay | Cells are treated for 72 h with inhibitors (including PU-WS13) or transfected with Grp94 siRNA or control siRNA, and their viability is assessed using CellTiter-Glo luminescent Cell Viability Assay. The method determines the number of viable cells in culture based on quantification of the ATP present, which signals the presence of metabolically active cells[1]. |
| References |
| Density | 1.5±0.1 g/cm3 |
|---|---|
| Boiling Point | 617.4±65.0 °C at 760 mmHg |
| Molecular Formula | C17H20Cl2N6S |
| Molecular Weight | 411.352 |
| Flash Point | 327.2±34.3 °C |
| Exact Mass | 410.084717 |
| PSA | 106.95000 |
| LogP | 4.00 |
| Vapour Pressure | 0.0±1.8 mmHg at 25°C |
| Index of Refraction | 1.703 |
| Storage condition | -20 ̊C |