| Structure | Name/CAS No. | Articles |
|---|---|---|
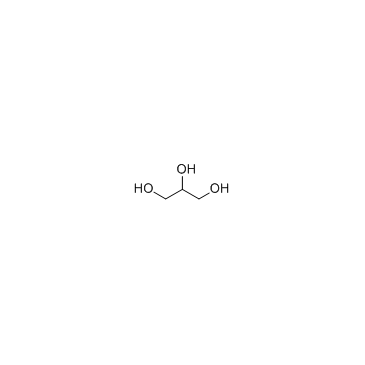 |
Glycerol
CAS:56-81-5 |
|
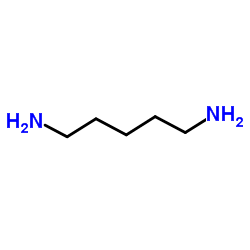 |
Cadaverine
CAS:462-94-2 |
|
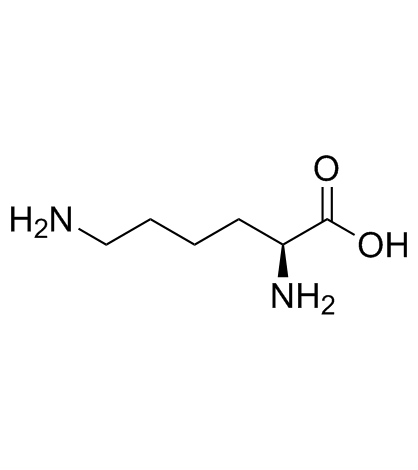 |
L-Lysine
CAS:56-87-1 |
|
 |
putrescine
CAS:110-60-1 |
|
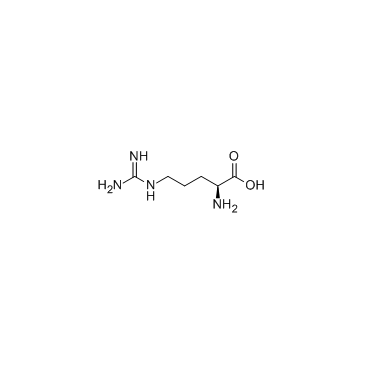 |
L-arginine
CAS:74-79-3 |