| Structure | Name/CAS No. | Articles |
|---|---|---|
 |
Sulfuric acid
CAS:7664-93-9 |
|
 |
Ethanol
CAS:64-17-5 |
|
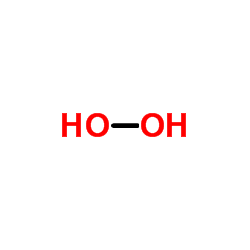 |
Hydrogen peroxide
CAS:7722-84-1 |
|
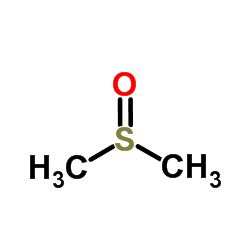 |
Dimethyl sulfoxide
CAS:67-68-5 |
|
 |
3-Aminopropyltriethoxysilane
CAS:919-30-2 |
|
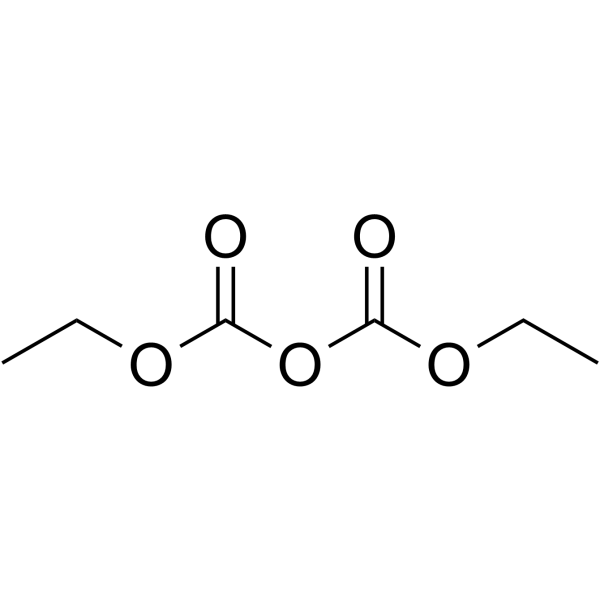 |
Diethyl pyrocarbonate
CAS:1609-47-8 |
|
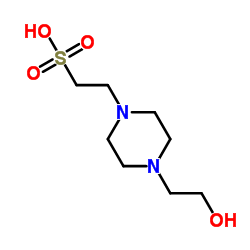 |
HEPES
CAS:7365-45-9 |
|
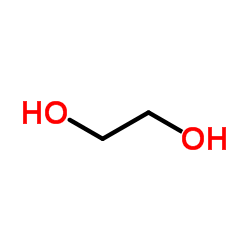 |
Ethylene glycol
CAS:107-21-1 |
|
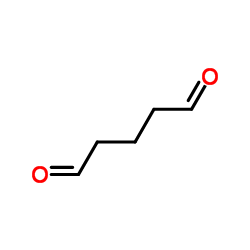 |
glutaraldehyde
CAS:111-30-8 |