| Structure | Name/CAS No. | Articles |
|---|---|---|
 |
sodium chloride
CAS:7647-14-5 |
|
 |
Glyceryl monooleate
CAS:25496-72-4 |
|
 |
SODIUM CHLORIDE-35 CL
CAS:20510-55-8 |
|
![4-[(3β)-Cholest-5-en-3-yloxy]-4-oxobutanoic acid Structure](https://image.chemsrc.com/caspic/456/1510-21-0.png) |
4-[(3β)-Cholest-5-en-3-yloxy]-4-oxobutanoic acid
CAS:1510-21-0 |
|
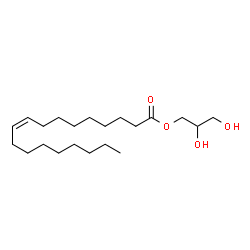 |
Monoolein
CAS:111-03-5 |
|
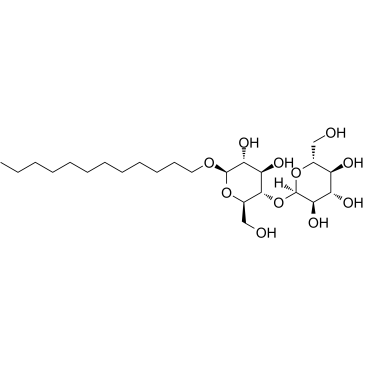 |
n-Dodecyl-beta-D-maltoside
CAS:69227-93-6 |