| Structure | Name/CAS No. | Articles |
|---|---|---|
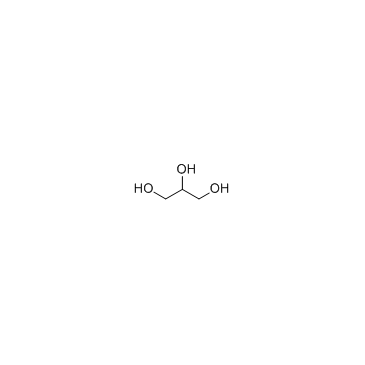 |
Glycerol
CAS:56-81-5 |
|
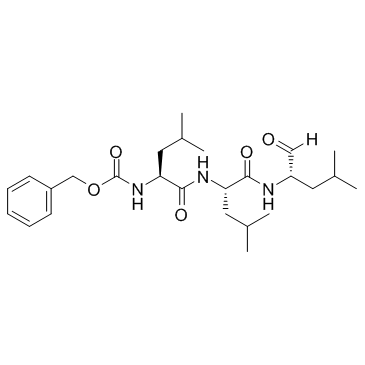 |
MG-132
CAS:133407-82-6 |
|
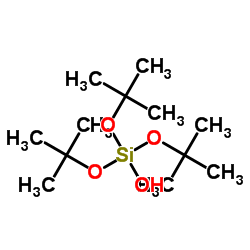 |
Tris(2-methyl-2-propanyl) hydrogen orthosilicate
CAS:18166-43-3 |
|
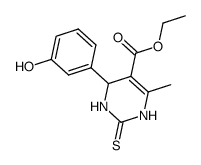 |
Monastrol
CAS:254753-54-3 |
|
 |
Nocodazole
CAS:31430-18-9 |