938440-64-3
| Name | Ku-0063794 |
|---|---|
| Synonyms |
KU-0063794
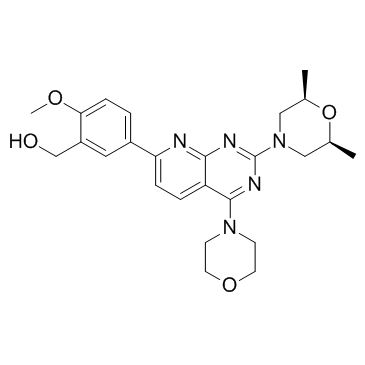
(5-{2-[(2R,6S)-2,6-Dimethyl-4-morpholinyl]-4-(4-morpholinyl)pyrido[2,3-d]pyrimidin-7-yl}-2-methoxyphenyl)methanol (5-{2-[(2R,6S)-2,6-dimethylmorpholin-4-yl]-4-(morpholin-4-yl)pyrido[2,3-d]pyrimidin-7-yl}-2-methoxyphenyl)methanol ku 0063794 KU0063794 |
| Description | KU-0063794 is a potent and specific mTOR inhibitor, inhibiting both the mTORC1 and mTORC2 complexes with IC50s of 10 nM. |
|---|---|
| Related Catalog | |
| Target |
mTORC1:10 nM (IC50) mTORC2:10 nM (IC50) |
| In Vitro | Ku-0063794 is cell permeant, suppresses activation and hydrophobic motif phosphorylation of Akt, S6K and SGK, but not RSK (ribosomal S6 kinase), an AGC kinase not regulated by mTOR. Ku-0063794 also inhibits phosphorylation of the T-loop Thr308 residue of Akt phosphorylated by PDK1 (3-phosphoinositide-dependent protein kinase-1). Ku-0063794 induces a much greater dephosphorylation of the mTORC1 substrate 4E-BP1 (eukaryotic initiation factor 4E-binding protein 1) than rapamycin, even in mTORC2-deficient cells, suggesting a form of mTOR distinct from mTORC1, or mTORC2 phosphorylates 4E-BP1. Ku-0063794 also suppresses cell growth and induced a G1-cell-cycle arrest[1]. Ku0063794 does not alter nuclear phospho-Mst1-Thr-120 levels in LNCaP cell nuclei, whereas Ku0063794 or CCI-779 increases phospho-Mst1-Thr-120 levels in C4-2 cell nuclei[2]. The combination of GDC-0941 and KU0063794 inhibits the phosphorylation of 4EBP1 and S6 to a similar extent to that caused by single agent NVP-BEZ235 in HCT116, DLD1 and HT29 cell lines[3]. |
| Kinase Assay | HEK-293 cells are freshly lysed in Hepes lysis buffer. Lysate (1-4 mg) is pre-cleared by incubating with 5-20 μL of Protein G-Sepharose conjugated to pre-immune IgG. The lysate extracts are then incubated with 5-20 μL of Protein G-Sepharose conjugated to 5-20 μg of either anti-Rictor or anti-Raptor antibody, or pre-immune IgG. All antibodies are covalently conjugated to Protein G-Sepharose. Immunoprecipitations are carried out for 1 hour at 4°C on a vibrating platform. The immunoprecipitates are washed four times with Hepes lysis buffer, followed by two washes with Hepes kinase buffer. For Raptor immunoprecipitates used for phosphorylating S6K1, for the initial two wash steps the buffer includes 0.5mol/LNaCl to ensure optimal kinase activity. GST-Akt1 is isolated from serum-deprived HEK-293 cells incubated with PI-103 (1 μM for 1 hour). GST-S6K1 is purified from serum-deprived HEK-293 cells incubated with rapamycin (0.1 μM for 1 hour). mTOR reactions are initiated by adding 0.1 mM ATP and 10 mM MgCl2 in the presence of various concentrations of KU-0063794 and GST-Akt1 (0.5 μg) or GST-S6K1 (0.5 μg). Reaction are carried out for 30 minutes at 30°C on a vibrating platform and stopped by addition of SDS sample buffer. Reaction mixtures are then filtered through a 0.22-μM-poresize Spin-X filter and samples are subjected to electrophoresis and immunoblot analysis with the indicated antibodies. |
| Cell Assay | Cells are treated with KU-0063794 for 24, 48, and 72 hours, and the medium is changed every 24 hours with freshly dissolved KU-0063794. For the measurement of cell growth, cells are washed once with PBS, and fixed in 4% (v/v) paraformaldehyde in PBS for 15 minutes. After washing once with water, the cells are stained with 0.1% Crystal Violet in 10% ethanol for 20 minutes and washed three times with water. Crystal Violet is extracted from cells with 0.5 mL of 10% (v/v) ethanoic (acetic) acid for 20 minutes. The eluate is then diluted 1:10 in water and absorbance at 590 nm is quantified. For the assessment of cell cycle distribution, cells are harvested by trypsinization, washed once in PBS, and re-suspended in ice-cold aq. 70% (v/v) ethanol. Cells are washed twice in PBS plus 1% (w/v) BSA and stained for 20 minutes in PBS plus 0.1% (v/v) Triton X-100 containing 50 g/mL propidium iodide and 50 g/mL RNase A. The DNA content of cells is determined using a FACSCalibur flow cytometer and CellQuest software. Red fluorescence (585 nm) is acquired on a linear scale, and pulse width analysis is used to exclude doublets. Cell-cycle distribution is determined using FlowJo software. |
| References |
| Density | 1.2±0.1 g/cm3 |
|---|---|
| Boiling Point | 694.3±65.0 °C at 760 mmHg |
| Molecular Formula | C25H31N5O4 |
| Molecular Weight | 465.545 |
| Flash Point | 373.7±34.3 °C |
| Exact Mass | 465.237610 |
| PSA | 93.07000 |
| LogP | 0.27 |
| Appearance | white to beige |
| Vapour Pressure | 0.0±2.3 mmHg at 25°C |
| Index of Refraction | 1.609 |
| Storage condition | Store at RT |
| Water Solubility | DMSO: >2mg/mL (warmed) |
| Symbol |

GHS06 |
|---|---|
| Signal Word | Danger |
| Hazard Statements | H301 |
| Precautionary Statements | P301 + P310 |
| Hazard Codes | T |
| Risk Phrases | 25 |
| Safety Phrases | 45 |
| RIDADR | UN 2811 6.1 / PGIII |