478182-28-4
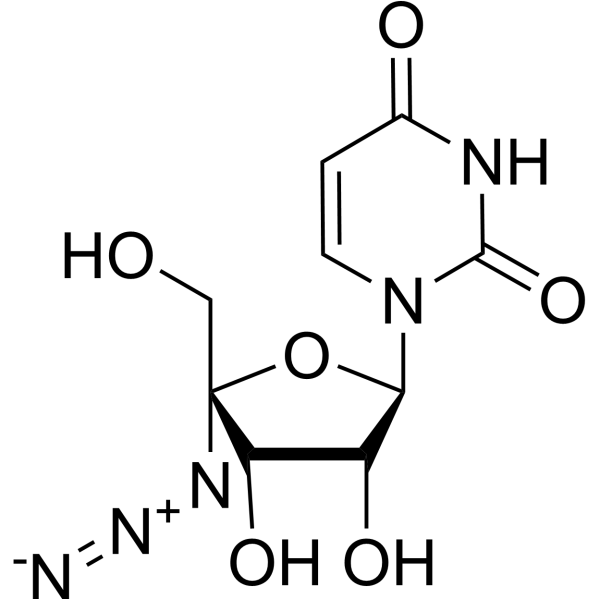
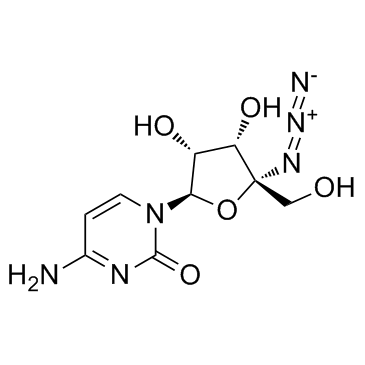
| Name | 4-amino-1-[(2R,3R,4S,5R)-5-azido-3,4-dihydroxy-5-(hydroxymethyl)oxolan-2-yl]pyrimidin-2-one |
|---|---|
| Synonyms |
4-amino-1-[(2R,3R,4S,5R)-5-azido-3,4-dihydroxy-5-(hydroxymethyl)tetrahydrofuran-2-yl]pyrimidin-2(1H)-one (non-preferred name)
R-1479||4'-Azidocytidine|R1479 CS-0362 4'-azidocytidine 4'azidocytidine R1479 4-Amino-1-[(2R,3R,4S,5R)-5-azido-3,4-dihydroxy-5-(hydroxymethyl)tetrahydro-2-furanyl]-2(1H)-pyrimidinone R-1479 |
| Description | R-1479 is a specific inhibitor of HCV replication in the HCV subgenomic replicon system (IC50=1.28 μM). |
|---|---|
| Related Catalog | |
| Target |
IC50: 1.28 μM (HCV replication)[1] |
| In Vitro | R-1479 (R1479) inhibits HCV RNA replication with a mean IC50 value of 1.28 μM when measured as dose-dependent reduction of Renillaluciferase activity after a 72 h incubation of proliferating replicon cells. R-1479 shows no effect on cell viability or proliferation of HCV replicon or Huh-7 cells at concentrations up to 2 mM[1]. The most potent and non-cytotoxic derivative is R-1479 with an IC50 of 1.28 μM in the HCV replicon system. The triphosphate of R-1479 is prepared and shown to be an inhibitor of RNA synthesis mediated by NS5B (IC50=320 nM), the RNA polymerase encoded by HCV. R-1479 displays good activity in the replicon assay with no measurable cytotoxic or cytostatic effect[2]. |
| Kinase Assay | The membrane-associated, native HCV replicase complex is isolated from 2209-23 HCV replicon cells and a derived cell line carrying HCV replicon RNA with a S282T mutation in the NS5B coding sequence. The in vitro replicase assay contain 10 μL of cytoplasmic membrane fraction, 50 mM HEPES (pH 7.5), 10 mM KCl, 10 mM dithiothreitol, 5 mM MgCl2, 20 μg/mL actinomycin D, 1 mM ATP, 1 mM GTP, 1 mM UTP, 30 μCi of [α-33P]CTP (3000 Ci/mmol, 10 mCi/mL), 1 unit/μL SUPERase•In, 10 mM creatine phosphate, and 200 μg/mL creatine phosphokinase in a final volume of 25 μL. Inhibition by nucleotide analogs is determined[1]. |
| Cell Assay | The effect of R-1479 on the incorporation of tritiated thymidine into cellular DNA is measured using the [3H]thymidine incorporation scintillation proximity assay system. MTT and WST-1 assay systems are used to measure cell viability. The ATP bioluminescence assay kit HSII is used to measure intracellular ATP levels[1]. |
| References |
| Molecular Formula | C9H12N6O5 |
|---|---|
| Molecular Weight | 284.229 |
| Exact Mass | 284.086914 |
| PSA | 181.57000 |
| LogP | -0.20 |
| Storage condition | -20°C |
|
~% 
478182-28-4 |
| Literature: Smith, David B.; Martin, Joseph A.; Klumpp, Klaus; Baker, Stewart J.; Blomgren, Peter A.; Devos, Rene; Granycome, Caroline; Hang, Julie; Hobbs, Christopher J.; Jiang, Wen-Rong; Laxton, Carl; Pogam, Sophie Le; Leveque, Vincent; Ma, Han; Maile, Graham; Merrett, John H.; Pichota, Arkadius; Sarma, Keshab; Smith, Mark; Swallow, Steven; Symons, Julian; Vesey, David; Najera, Isabel; Cammack, Nick Bioorganic and Medicinal Chemistry Letters, 2007 , vol. 17, # 9 p. 2570 - 2576 |
|
~% 
478182-28-4 |
| Literature: Smith, David B.; Martin, Joseph A.; Klumpp, Klaus; Baker, Stewart J.; Blomgren, Peter A.; Devos, Rene; Granycome, Caroline; Hang, Julie; Hobbs, Christopher J.; Jiang, Wen-Rong; Laxton, Carl; Pogam, Sophie Le; Leveque, Vincent; Ma, Han; Maile, Graham; Merrett, John H.; Pichota, Arkadius; Sarma, Keshab; Smith, Mark; Swallow, Steven; Symons, Julian; Vesey, David; Najera, Isabel; Cammack, Nick Bioorganic and Medicinal Chemistry Letters, 2007 , vol. 17, # 9 p. 2570 - 2576 |
|
~% 
478182-28-4 |
| Literature: Smith, David B.; Martin, Joseph A.; Klumpp, Klaus; Baker, Stewart J.; Blomgren, Peter A.; Devos, Rene; Granycome, Caroline; Hang, Julie; Hobbs, Christopher J.; Jiang, Wen-Rong; Laxton, Carl; Pogam, Sophie Le; Leveque, Vincent; Ma, Han; Maile, Graham; Merrett, John H.; Pichota, Arkadius; Sarma, Keshab; Smith, Mark; Swallow, Steven; Symons, Julian; Vesey, David; Najera, Isabel; Cammack, Nick Bioorganic and Medicinal Chemistry Letters, 2007 , vol. 17, # 9 p. 2570 - 2576 |
|
~% 
478182-28-4 |
| Literature: Smith, David B.; Martin, Joseph A.; Klumpp, Klaus; Baker, Stewart J.; Blomgren, Peter A.; Devos, Rene; Granycome, Caroline; Hang, Julie; Hobbs, Christopher J.; Jiang, Wen-Rong; Laxton, Carl; Pogam, Sophie Le; Leveque, Vincent; Ma, Han; Maile, Graham; Merrett, John H.; Pichota, Arkadius; Sarma, Keshab; Smith, Mark; Swallow, Steven; Symons, Julian; Vesey, David; Najera, Isabel; Cammack, Nick Bioorganic and Medicinal Chemistry Letters, 2007 , vol. 17, # 9 p. 2570 - 2576 |
|
~% 
478182-28-4 |
| Literature: Smith, David B.; Martin, Joseph A.; Klumpp, Klaus; Baker, Stewart J.; Blomgren, Peter A.; Devos, Rene; Granycome, Caroline; Hang, Julie; Hobbs, Christopher J.; Jiang, Wen-Rong; Laxton, Carl; Pogam, Sophie Le; Leveque, Vincent; Ma, Han; Maile, Graham; Merrett, John H.; Pichota, Arkadius; Sarma, Keshab; Smith, Mark; Swallow, Steven; Symons, Julian; Vesey, David; Najera, Isabel; Cammack, Nick Bioorganic and Medicinal Chemistry Letters, 2007 , vol. 17, # 9 p. 2570 - 2576 |
|
~% 
478182-28-4 |
| Literature: Smith, David B.; Martin, Joseph A.; Klumpp, Klaus; Baker, Stewart J.; Blomgren, Peter A.; Devos, Rene; Granycome, Caroline; Hang, Julie; Hobbs, Christopher J.; Jiang, Wen-Rong; Laxton, Carl; Pogam, Sophie Le; Leveque, Vincent; Ma, Han; Maile, Graham; Merrett, John H.; Pichota, Arkadius; Sarma, Keshab; Smith, Mark; Swallow, Steven; Symons, Julian; Vesey, David; Najera, Isabel; Cammack, Nick Bioorganic and Medicinal Chemistry Letters, 2007 , vol. 17, # 9 p. 2570 - 2576 |
|
~% 
478182-28-4 |
| Literature: Smith, David B.; Martin, Joseph A.; Klumpp, Klaus; Baker, Stewart J.; Blomgren, Peter A.; Devos, Rene; Granycome, Caroline; Hang, Julie; Hobbs, Christopher J.; Jiang, Wen-Rong; Laxton, Carl; Pogam, Sophie Le; Leveque, Vincent; Ma, Han; Maile, Graham; Merrett, John H.; Pichota, Arkadius; Sarma, Keshab; Smith, Mark; Swallow, Steven; Symons, Julian; Vesey, David; Najera, Isabel; Cammack, Nick Bioorganic and Medicinal Chemistry Letters, 2007 , vol. 17, # 9 p. 2570 - 2576 |
| Precursor 3 | |
|---|---|
| DownStream 0 | |