| Description |
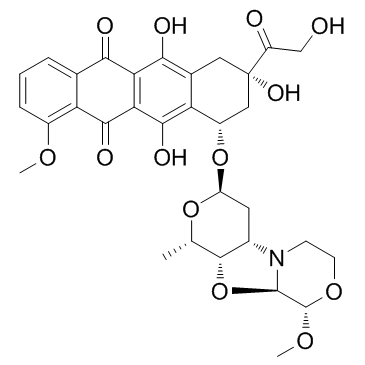
PNU-159682, a highly potent metabolite of the anthracycline nemorubicin with outstanding cytotoxicity, is a potent ADCs cytotoxin.
|
| Related Catalog |
|
| In Vitro |
PNU-159682 inhibits a panel of human tumor cell lines with IC70 values in the range of 0.07-0.58 nM, and is 2,360- to 790-fold and 6,420- to 2,100-fold more potent than MMDX and doxorubicin, respectively[1]. PNU-159682 (100 μM) weakly inhibits topoisomerase II unknotting activity. PNU-159682 (10 μM)-DNA adducts contain one or two drug molecules bound to double-stranded DNA[2]. PNU-159682 shows cytotoxic effect on CAIX-expressing SKRC-52 cells with IC50 of 25 nM[3].
|
| In Vivo |
PNU-159682 (15 μg/kg, i.v.) shows antitumor activity in mice bearing disseminated murine L1210 leukemia and in MX-1 human mammary carcinoma xenografts at 4 μg/kg[1]. PNU-159682 (25 nmol/kg) exhibits a potent antitumor effect in mice bearing SKRC-52 xenografted tumors[3].
|
| Kinase Assay |
The inhibition of topoisomerase II activity is tested by taking advantage of the ability of this enzyme to decatenate kinetoplast DNA (kDNA); the test is specific for both isoforms of topoisomerase II (α and β) because it relies on the conversion of catenated DNA to its decatenated form, which requires double strand cut and ligation uniquely performed by topoisomerase II. The DNA used in this test is the mitochondrial kDNA of Crithidia fasciculata, a catenated network of DNA rings, most of which are 2.5 kb monomers. The kDNA networks are large relative to the monomers and do not migrate in the gel remaining in the well, while the minicircles can be easily resolved in agarose gel. Both the gel and the running buffer contain the intercalator ethidium bromide, which allows the monitoring of monomers appearance with a UV light source and the resolution of different DNA forms (linear, nicked circular DNA, and relaxed DNA monomers), helping to clearly distinguish the linear DNA from the nicked circular DNA. In this test, 200 ng of kDNAs is incubated for 1 h at 37°C with doxorubicin at 10, 1, or 0.1 μM, or with PNU-159682 at 100, 10, or 1 μM in the presence of 0.025 U of human topoisomerase IIα in a topoisomerase II reaction buffer (Tris-HCl pH 7.9 40 mM, KCl 80 mM, DTT 5 mM, BSA 15 μg/mL, ATP 1 mM, and MgCl2 10 mM). At the end of the incubation period, each sample is spiked with 3 μL of gel loading buffer (xylene cyanol 0.25%, blue bromophenol 0.25%, Ficoll 400 18%, and EDTA 6 mM) and then analyzed by agarose (1%) gel electrophoresis. Runs are performed in TBE buffer (Tris 89 mM, boric Acid 89 mM, EDTA 2 mM, pH 8.0) in the presence of ethidium bromide 0.5 μg/mL. Samples are run overnight at 1 V/cm.
|
| Cell Assay |
SKRC-52 cells are seeded in 96-well plates in RPMI added with 10% FCS (100 μL) at a density of 5000 cells per well and allowed to grow for 24 h. The medium is replaced with medium containing different concentrations of test substance (1:3 dilution steps) and plates are incubated under standard culture conditions. After 72 h the medium is removed, MTS cell viability dye (20 μL) in 150 μL of the medium is added, the plates are incubated for 2 h under culture conditions and the absorbance at 490 nm measured on a Spectra Max Paradigm multimode plate reader. Experiments are performed in triplicate and average cell viability calculated as measured background corrected absorbance divided by the absorbance of untreated control wells. IC50 values are determined by fitting data to the four-parameter logistic equation, using a Prism 6 software for data analysis.
|
| Animal Admin |
Four- to six-week-old female CD-1 athymic nude mice are used for evaluation of the activity of PNU-159682 against MX-1 human mammary carcinoma xenografts. On day 0, animals (n=14) are grafted s.c. with MX-1 tumor fragments in the right flank. Eight days later, they are randomly assigned to the drug treatment group or control group (n=7 mice per group), and treatment is started. PNU-159682 is given i.v. (4 μg/kg) according to a q7dx3 (every 7 days for three doses) schedule; control animals receive saline injections. Tumor volume is estimated from measurements done with a caliper; where D and d are the longest and the shortest diameters, respectively. For ethical reasons, control animals are sacrificed on day 21 when the mean tumor volume in the group is appr 2,500 mm3; animals receiving drug treatment are monitored up to day 50, at which point they are sacrificed.
|
| References |
[1]. Quintieri L, et al. Formation and antitumor activity of PNU-159682, a major metabolite of nemorubicin in human liver microsomes. Clin Cancer Res. 2005 Feb 15;11(4):1608-17. [2]. Scalabrin M, et al. Virtual Cross-Linking of the Active Nemorubicin Metabolite PNU-159682 to Double-Stranded DNA. Chem Res Toxicol. 2017 Feb 20;30(2):614-624. [3]. Cazzamalli S, et al. Acetazolamide Serves as Selective Delivery Vehicle for Dipeptide-Linked Drugs to Renal Cell Carcinoma. Mol Cancer Ther. 2016 Dec;15(12):2926-2935.
|