
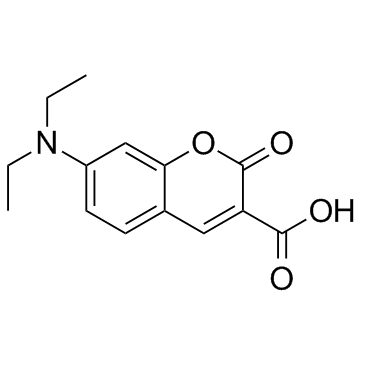
7-(二乙氨基)香豆素-3-甲酸结构式

|
常用名 | 7-(二乙氨基)香豆素-3-甲酸 | 英文名 | 7-Diethylaminocoumarin-3-carboxylic acid |
|---|---|---|---|---|
| CAS号 | 50995-74-9 | 分子量 | 261.273 | |
| 密度 | 1.3±0.1 g/cm3 | 沸点 | 464.7±45.0 °C at 760 mmHg | |
| 分子式 | C14H15NO4 | 熔点 | 222-224ºC (dec.)(lit.) | |
| MSDS | 中文版 美版 | 闪点 | 234.9±28.7 °C |
|
Photophysical properties of 7-(diethylamino)coumarin-3-carboxylic acid in the nanocage of cyclodextrins and in different solvents and solvent mixtures.
Photochem. Photobiol. , (2012) The photophysical properties of 7-(diethylamino) coumarin-3-carboxylic acid (7-DCCA) were studied in cyclodextrins (α, β, γ,-CDs), different neat solvents and solvent mixtures by using steady state absorption, emission and time-resolved fluorescence spectrosc... |
|
|
Cholesterol-dependent formation of GM1 ganglioside-bound amyloid beta-protein, an endogenous seed for Alzheimer amyloid.
J. Biol. Chem. 276(27) , 24985-90, (2001) GM1 ganglioside-bound amyloid beta-protein (GM1/Abeta), found in brains exhibiting early pathological changes of Alzheimer's disease (AD) including diffuse plaques, has been suggested to be involved in the initiation of amyloid fibril formation in vivo by act... |
|
|
Interactions of amyloid beta-protein with various gangliosides in raft-like membranes: importance of GM1 ganglioside-bound form as an endogenous seed for Alzheimer amyloid.
Biochemistry 41(23) , 7385-90, (2002) GM1 ganglioside-bound amyloid beta-protein (GM1-Abeta), found in brains exhibiting early pathological changes of Alzheimer's disease (AD) plaques, has been suggested to accelerate amyloid fibril formation by acting as a seed. We have previously found using dy... |
|
|
Inhibition of the formation of amyloid beta-protein fibrils using biocompatible nanogels as artificial chaperones.
FEBS Lett. 580(28-29) , 6587-95, (2006) The formation of fibrils by amyloid beta-protein (Abeta) is considered as a key step in the pathology of Alzheimer's disease (AD). Inhibiting the aggregation of Abeta is a promising approach for AD therapy. In this study, we used biocompatible nanogels compos... |
|
|
Driving force of binding of amyloid beta-protein to lipid bilayers.
Biochem. Biophys. Res. Commun. 370(3) , 525-9, (2008) Amyloid beta-protein (Abeta) has been reported to interact with a variety of lipid species, although the thermodynamic driving force remains unclear. We investigated the binding of Abetas labeled with the dye diethylaminocoumarin (DAC-Abetas) to lipid bilayer... |
|
|
Crystal structure and photoluminescence of 7-(N,N'-diethylamino)-coumarin-3-carboxylic acid.
Spectrochim. Acta. A. Mol. Biomol. Spectrosc. 69(4) , 1136-9, (2008) The structure of 7-(N,N'-diethylamino)-coumarin-3-carboxylic acid (DCCA) was verified by single crystal X-ray crystallography. The UV-vis absorption and fluorescence of DCCA were discussed. The compound exhibits strong blue emission under ultraviolet light ex... |
|
|
Quantitation of myoglobin saturation in the perfused heart using myoglobin as an optical inner filter.
Am. J. Physiol. 267(2 Pt 2) , H645-53, (1994) Quantitation of metabolic parameters using the technique of cardiac surface fluorescence is complicated by motion and changes in tissue absorption. Because ratio fluorescence methodology can be applied to eliminate motion-induced errors, in the current study,... |
|
|
Mechanism of interaction between single-stranded DNA binding protein and DNA.
Biochemistry 49(5) , 843-52, (2010) A single-stranded DNA binding protein (SSB), labeled with a fluorophore, interacts with single-stranded DNA (ssDNA), giving a 6-fold increase in fluorescence. The labeled protein is the adduct of the G26C mutant of the homotetrameric SSB from Escherichia coli... |
|
|
Design of irreversible optical nanothermometers for thermal ablations.
Chem. Commun. (Camb.) 49(7) , 680-2, (2013) Nanothermometers composed from a gold nanorod core, temperature sensitive linker and fluorescent dye are reported. The nanothermometers have low fluorescence due to a self-quenching mechanism at temperatures below 50 °C and become highly fluorescence above 70... |
|
|
Intense blue fluorescence in a leucine zipper assembly.
ChemBioChem. 12(5) , 691-4, (2011)
|