| Structure | Name/CAS No. | Articles |
|---|---|---|
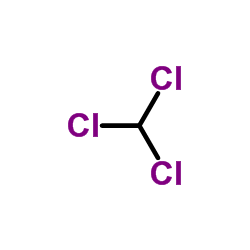 |
Chloroform
CAS:67-66-3 |
|
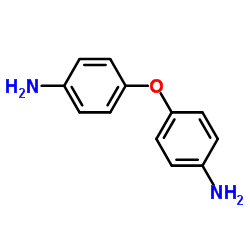 |
4,4'-Oxydianiline
CAS:101-80-4 |
|
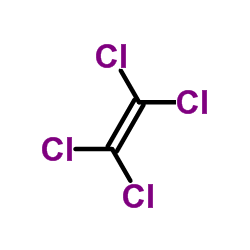 |
Tetrachloroethylene
CAS:127-18-4 |
|
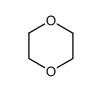 |
1,4-Dioxane
CAS:123-91-1 |
|
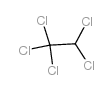 |
Ethane,1,1,1,2,2-pentachloro
CAS:76-01-7 |
|
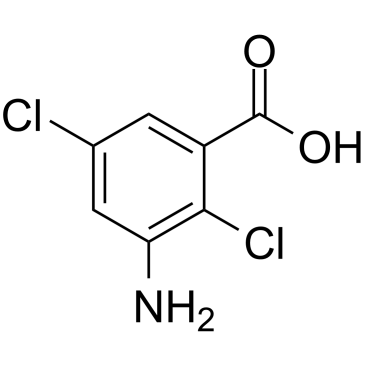 |
chloramben
CAS:133-90-4 |
|
 |
Dichloromethane
CAS:75-09-2 |
|
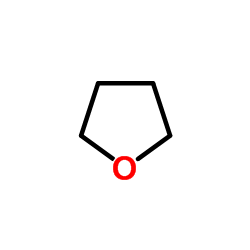 |
thf
CAS:109-99-9 |
|
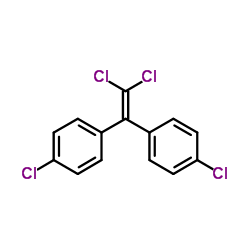 |
p,p'-DDE
CAS:72-55-9 |
|
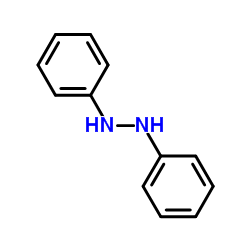 |
1,2-Diphenylhydrazine
CAS:122-66-7 |