| Structure | Name/CAS No. | Articles |
|---|---|---|
 |
sodium chloride
CAS:7647-14-5 |
|
 |
Methanol
CAS:67-56-1 |
|
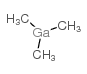 |
Trimethyl-Gallium
CAS:1445-79-0 |
|
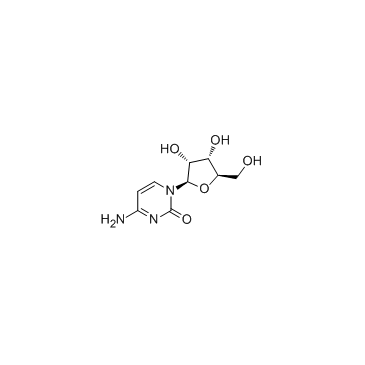 |
Cytidine
CAS:65-46-3 |
|
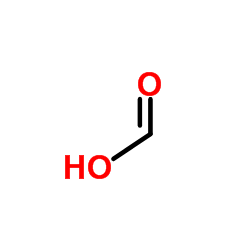 |
Formic Acid
CAS:64-18-6 |
|
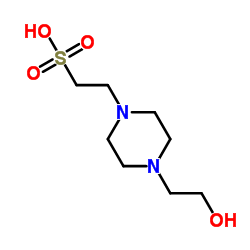 |
HEPES
CAS:7365-45-9 |
|
 |
SODIUM CHLORIDE-35 CL
CAS:20510-55-8 |
|
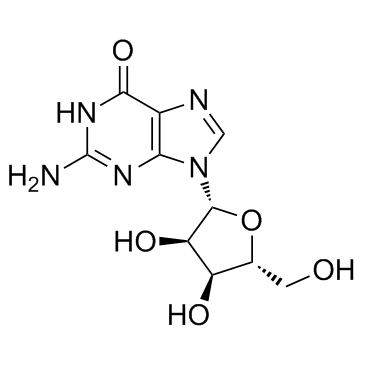 |
Guanosine
CAS:118-00-3 |
|
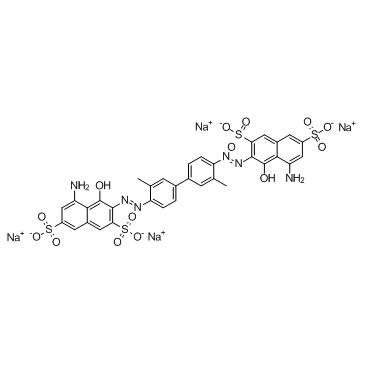 |
Direct Blue 14
CAS:72-57-1 |
|
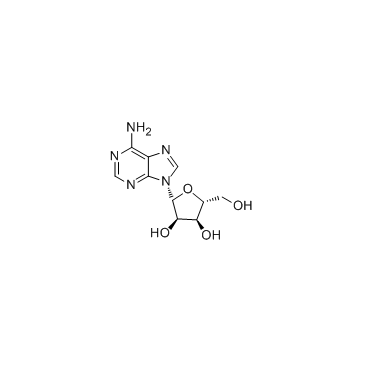 |
Adenosine
CAS:58-61-7 |