| Structure | Name/CAS No. | Articles |
|---|---|---|
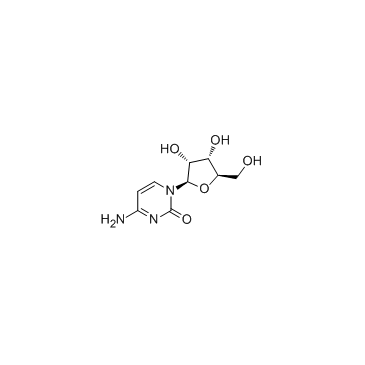 |
Cytidine
CAS:65-46-3 |
|
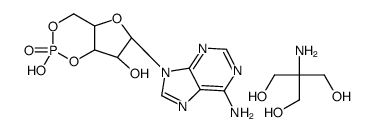 |
ADENOSINE 3':5'-CYCLIC MONOPHOSPHATE TRIS SALT
CAS:102029-77-6 |
|
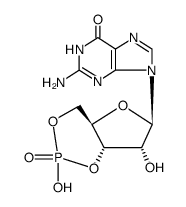 |
Guanosine 3',5'-cyclic monophosphate
CAS:7665-99-8 |
|
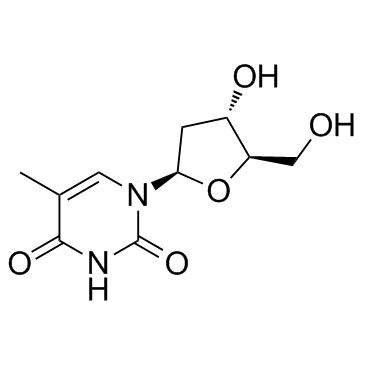 |
Thymidine
CAS:50-89-5 |
|
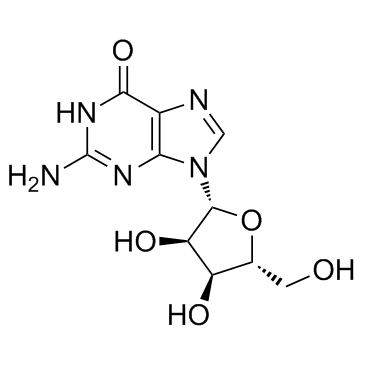 |
Guanosine
CAS:118-00-3 |
|
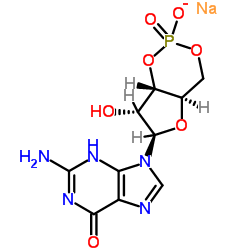 |
guanosine 3':5'-cyclic monophosphate sodium salt
CAS:40732-48-7 |
|
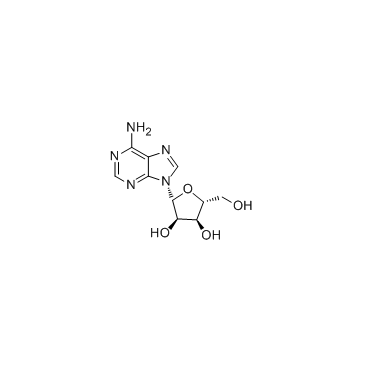 |
Adenosine
CAS:58-61-7 |
|
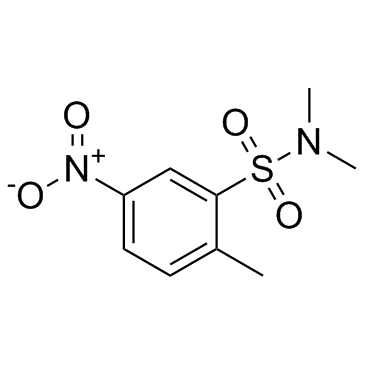 |
BRL-50481
CAS:433695-36-4 |
|
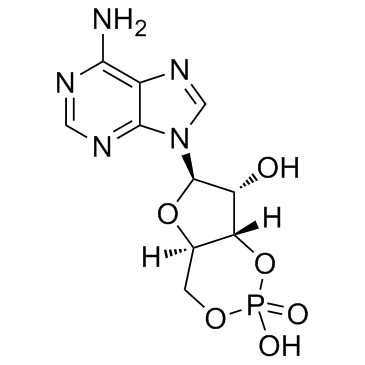 |
Adenosine cyclophosphate
CAS:60-92-4 |
|
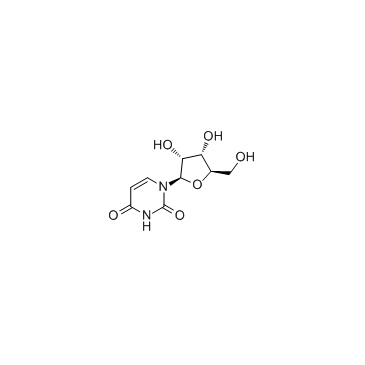 |
Uridine
CAS:58-96-8 |