| In Vitro |
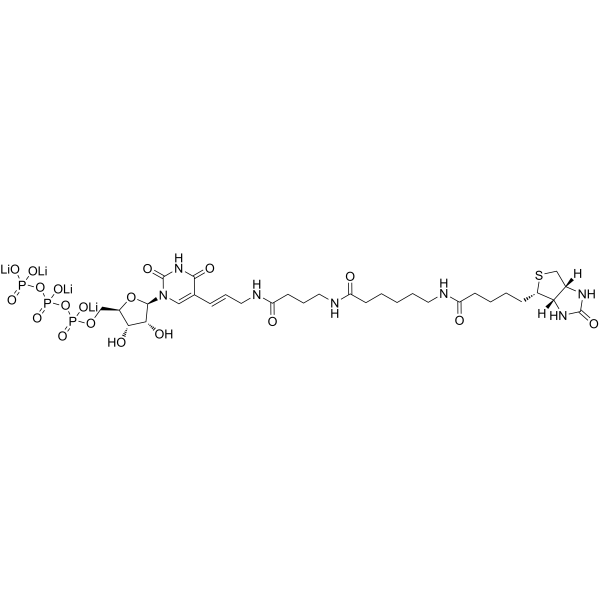
Guidelines (Following is our recommended protocol. This protocol only provides a guideline, and should be modified according to your specific needs). In Vitro RNA Synthesis and Purification: 1. Incubate the cells according to your normal protocol. 2. Add gently One volume of transcription buffer 2x [200 mM KCI, 20 mM Tris-HCI, pH 8.0, 5 mM MgCl2, 4 mM dithiothreitol (DTT), 4 mM each of ATP, GTP and CTP, 200 mM sucrose and 20% glycerol] to nuclei in ice, form mixture. 3. Add 8 μL biotin-16-UTP (from 10 mM tetralithium sal) to the mixture, which is incubated for 30 min at 29°C. 4. Add 6 μL 250 mM CaCl2, 6 μL RNase-free DNase I (10 U/μL) and incubating for 10 min at 29°C to stop reaction. 5. Perform RNA purification of both nuclear run-on and total RNA according to the manufacturer's instructions. 6. Resuspend RNA in 50 uL diethylpyrocarbonate (DEPC)-treated water. 7. Labeled RNA was captured by streptavidin-coated magnetic beads. 8. RNA-binding beads are then used for random hexamer primed reverse transcription.
|