L-SERINE BETA-NAPHTHYLAMIDE
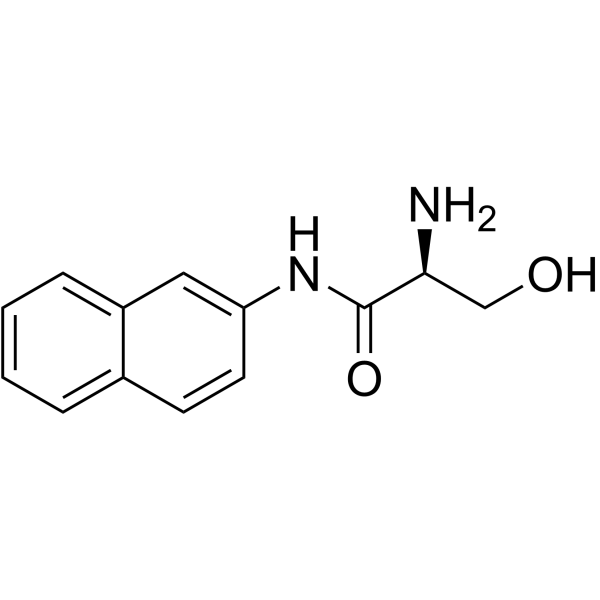
L-SERINE BETA-NAPHTHYLAMIDE structure
|
Common Name | L-SERINE BETA-NAPHTHYLAMIDE | ||
|---|---|---|---|---|
| CAS Number | 888-74-4 | Molecular Weight | 230.26200 | |
| Density | N/A | Boiling Point | N/A | |
| Molecular Formula | C13H14N2O2 | Melting Point | N/A | |
| MSDS | Chinese USA | Flash Point | N/A | |
| Symbol |

GHS08 |
Signal Word | Warning | |
|
Purification and characterization of a Cl- -activated aminopeptidase from bovine skeletal muscle.
Biosci. Biotechnol. Biochem. 70 , 1110-1117, (2006) To elucidate the mechanisms involved in the increase in free amino acids during postmortem storage of meat, a novel aminopeptidase was purified from bovine skeletal muscle by ammonium sulfate fractionation and successive chromatographies such as DEAE-cellulos... |
|
|
Gene cloning and characterization of PepC, a cysteine aminopeptidase from Streptococcus thermophilus, with sequence similarity to the eucaryotic bleomycin hydrolase.
Eur. J. Biochem. 224 , 497-506, (1994) Streptococcus thermophilus CNRZ 302 contains at least three general aminopeptidases able to hydrolyze Phe-beta-naphthylamide substrate. The gene encoding one of these aminopeptidases was cloned from a total DNA library of S. thermophilus CNRZ 302 constructed ... |
|
|
Molecular cloning and DNA sequence analysis of pepL, a leucyl aminopeptidase gene from Lactobacillus delbrueckii subsp. lactis DSM7290.
Eur. J. Biochem. 228 , 570-578, (1995) A genomic library of Lactobacillus delbrueckii subsp. lactis DSM7290 DNA fragments from a Sau3A partial digestion in the low-copy-number vector pLG339, was used to screen Escherichia coli for the presence of peptidases. Using the chromogenic substrate leucine... |
|
|
Proteolytic cleavage of the puromycin-sensitive aminopeptidase generates a substrate binding domain.
Arch. Biochem. Biophys. 415 , 80-86, (2003) The puromycin-sensitive aminopeptidase was found to be resistant to proteolysis by trypsin, chymotrypsin, and protease V8 but was cleaved into an N-terminal 60-kDa fragment and a C-terminal 33-kDa fragment by proteinase K. The two proteinase K fragments remai... |