SCR7 pyrazine
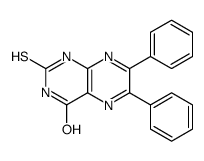
SCR7 pyrazine structure
|
Common Name | SCR7 pyrazine | ||
|---|---|---|---|---|
| CAS Number | 14892-97-8 | Molecular Weight | 332.37900 | |
| Density | N/A | Boiling Point | N/A | |
| Molecular Formula | C18H12N4OS | Melting Point | 209 °C | |
| MSDS | Chinese USA | Flash Point | N/A | |
| Symbol |

GHS07 |
Signal Word | Warning | |
|
Increasing the efficiency of CRISPR/Cas9-mediated precise genome editing in rats by inhibiting NHEJ and using Cas9 protein.
.PubMed ID Precise modifications such as site mutation, codon replacement, insertion or precise targeted deletion are needed for studies of accurate gene function. The CRISPR/Cas9 system has been proved as a powerful tool to generate gene knockout and knockin animals. B... |
|
|
Enrichment of G2/M cell cycle phase in human pluripotent stem cells enhances HDR-mediated gene repair with customizable endonucleases.
Sci. Rep. 6 , 21264, (2016) Efficient gene editing is essential to fully utilize human pluripotent stem cells (hPSCs) in regenerative medicine. Custom endonuclease-based gene targeting involves two mechanisms of DNA repair: homology directed repair (HDR) and non-homologous end joining (... |
|
|
Increasing the efficiency of precise genome editing with CRISPR-Cas9 by inhibition of nonhomologous end joining.
Nat. Biotechnol. 33(5) , 538-42, (2015) Methods to introduce targeted double-strand breaks (DSBs) into DNA enable precise genome editing by increasing the rate at which externally supplied DNA fragments are incorporated into the genome through homologous recombination. The efficiency of these metho... |
|
|
An inhibitor of nonhomologous end-joining abrogates double-strand break repair and impedes cancer progression.
Cell 151(7) , 1474-87, (2012) DNA Ligase IV is responsible for sealing of double-strand breaks (DSBs) during nonhomologous end-joining (NHEJ). Inhibiting Ligase IV could result in amassing of DSBs, thereby serving as a strategy toward treatment of cancer. Here, we identify a molecule, SCR... |