| Structure | Name/CAS No. | Articles |
|---|---|---|
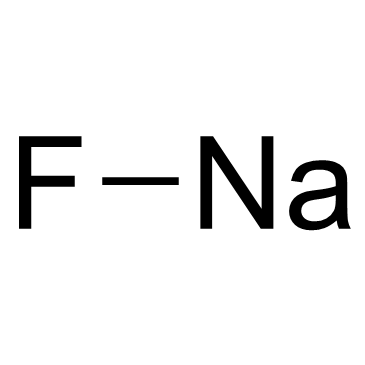 |
Sodium Fluoride
CAS:7681-49-4 |
|
 |
sodium chloride
CAS:7647-14-5 |
|
 |
L-γ-Glutamyl-S-nitroso-L-cysteinylglycine
CAS:57564-91-7 |
|
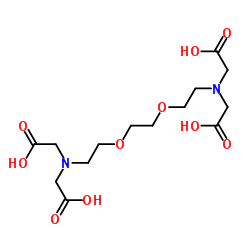 |
EGTA
CAS:67-42-5 |
|
 |
SODIUM CHLORIDE-35 CL
CAS:20510-55-8 |
|
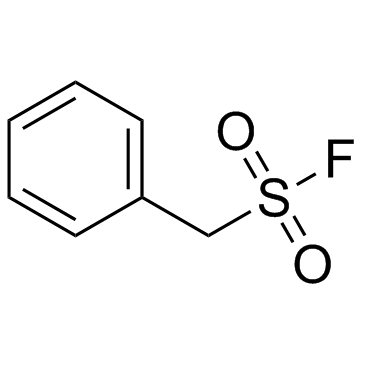 |
PMSF
CAS:329-98-6 |
|
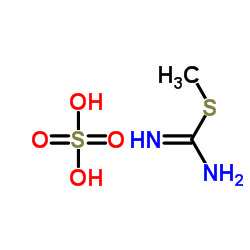 |
S-Methylisothiourea sulfate
CAS:867-44-7 |
|
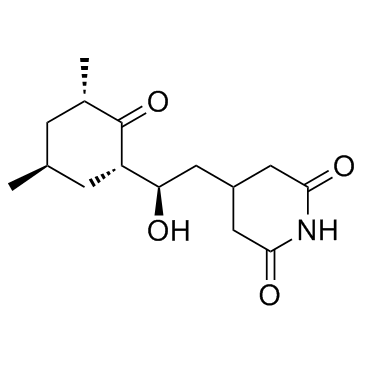 |
Cycloheximide
CAS:66-81-9 |
|
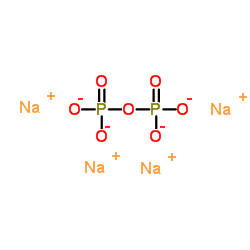 |
Tetrasodium pyrophosphate
CAS:7722-88-5 |